Wastewater Treatment Plants; Hot Spots of Emerging Contaminants in the Environment
| Received 07 Dec, 2022 |
Accepted 27 May, 2023 |
Published 07 Jun, 2023 |
Background and Objective: The Wupa wastewater treatment plant is a major sewage treatment plant in Abuja, Nigeria’s capital. While, the Shahe Reservoir treatment plant is a major treatment plant in Beijing, China’s capital. In this study, the quantification of selected antibiotics, the occurrence of antibiotic resistance genes and physicochemical parameters were assayed for both study areas. Materials and Methods: Samples from both treatment plants were collected for physicochemical and molecular studies. Samples were analyzed for total nitrogen (TN), total phosphorus (TP), chemical oxygen demand (COD) and total chemical oxygen demand (TCOD). Antibiotic residues were detected by the Solid Phase Extraction and Liquid Chromatography Method (SPE-LM). While antibiotics resistance genes were determined by Polymerase Chain Reaction (PCR). Results: About seventeen different antibiotics were detected with Ciprofloxacin having the highest concentration. Antibiotic resistance genes, Beta-lactam (blaOXA-1), Sulfonamide resistance genes (sul1 and sul2) and mobile genetic element, Class1 integron (Int1) were amplified from both study areas. Conclusion: Antibiotics and antibiotic resistance genes were confirmed in the study. The intl-1 gene, amplified for samples from both study areas may facilitate the proliferation and propagation of some antibiotic-resistant genes ARGs in this environment and represents public health risks. The need for control of potential ARG contamination is recommended.
| Copyright © 2023 Abakpa et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Water shortages remain among the most pressing challenges, the world currently faces1. As the human population continues to grow and urbanize, the challenges of securing water resources and disposing of wastewater become increasingly more difficult2. Developing countries of which Nigeria is one are disproportionately affected by a lack of access to clean water. China, the second largest economic entity in the world, is confronted by severe water issues as a result of its rapid expansion and urbanization transition3. Water reuse offers tremendous potential in augmenting already strained water resource portfolios. However, emerging contaminants such as pharmaceuticals and antibiotic-resistant bacteria are a source of concern. Increasing acquired antibiotic resistance is among the greatest worldwide concerns for health care and is currently considered one of the most serious public health issues. A large number of antibiotics are being used as precaution and therapy in industrial high-density animal farms worldwide4. It was reported that about 10 million people will die from antimicrobial resistance, AMR, annually by 2050 if the current trend continues, 40% of these deaths are proposed to be from Africa5. Antibiotic-resistant bacteria (ARB) can survive the inhibitory action of one or more antibiotics6.
China is the largest producer and consumer of antibiotics in the world, accounting for approximately 72,700 tons of 36 common antibiotics consumed in 2013, divided between clinical and veterinary use7, which all end up as waste in water bodies. These water bodies are sources of water reuse. Wastewater reuse may have implications at two different levels, alter the physicochemical and microbiological properties of the soil and/or introduce and contribute to the chemical and biological contaminants in the soil. These affect soil productivity and fertility and pose serious risks to human and environmental health. Previously, fingerprints of resistance Escherichia coli O157:H7 from vegetables and environmental samples from wastewater reuse in irrigation in Nigeria were reported8. Sustainable wastewater reuse should prevent both types of effects, requiring a holistic and integrated risk assessment9.
This study aimed at an assessment and determination of the presence of some selected commonly used antibiotics and antibiotics resistance genes in wastewater samples from two treatment plants in Nigeria and Beijing respectively with the following objectives, determination of some physicochemical parameters of the water samples (TN, TP, COD and TCO), determination of antibiotics by SPE-LM, determination of some antibiotic resistance genes by PCR.
MATERIALS AND METHODS
Study area: Samples were obtained from two study areas from July to September, 2018. The Wupa wastewater treatment plant in Abuja, Nigeria and the Shahe Reservoir in Beijing China. The Wupa Sewage Treatment Plant is located within the Idu-Industrial Area of Abuja Municipal on coordinates 7°23"N, 9°01"E next to the Wupa River where effluents from the Wupa sewage treatment plant discharges. It covers an area of 297,900 m2 with surrounding rural settlements. Residents of the Wupa village located downstream of the treatment plant use the river to meet their domestic water needs. While the Shahe Reservoir upstream of Beijing is a shallow lake from where the Wenyu River (called the North Canal further downstream) runs for 240 km to Bohai Bay. It receives a large fraction of the Beijing wastewater treatment plant effluents. Two major canals divert water from the North Canal for irrigation to the agricultural area between Beijing and Tianjin. The Beijing-Tianjin Region has the highest growth rates in economy and population and therefore, faces increasingly severe water scarcity10.
Research protocol: A total of 32 wastewater samples were analyzed in the study comprising 10 samples from the Wupa wastewater treatment plant in Abuja-Nigeria and 22 from the Shahe reservoir in Beijing, China. Samples were analyzed for some physicochemical parameters using a UV spectrophotometer. The presence of antibiotic residues was detected by SPE-LM. While antibiotics resistance genes were determined by PCR. Antibiotic resistance gene markers assayed include, blaOXA-1, Sul1, Sul2 and Int1 gene using their specific primers.
Sample pretreatment and DNA extraction: Water samples (500 mL) were filtered through 0.2 μm pore size polycarbonate membranes (GTTP, Millipore, Ireland) and the filter membranes were cut into small pieces and placed in extraction tubes provided in DNA Kit (Omega, USA). Total DNA was extracted according to the instructions of the manufacturer. The quality of extraction was verified by agarose gel electrophoresis and the DNA quantities were measured using a UV-Vis spectrophotometer (Q5000, Quawell, USA). Amplification was performed using a thermal cycler, in Cialifornia, USA. A negative control (reaction tube with nuclease-free water) was included as quality control. Sequences of all primers of the target genes and PCR conditions were given in Table 1.
| Table 1: | Primer sequences, PCR preparations and conditions used in the study | |||
| Target | Primer sequence | PCR preparation (25 μL) | PCR conditions |
| sul1 | Sul1 F | 12.5 μL Dream Taq master mix, | 3 min at 95°C before 35 cycles of 1 min |
| 5'-CCGTTGGCCTTCCTGTAAAG -3' | 8.5 μL uclease-free water, | at 94°C, annealing temperature at 47°C | |
| Sul1 R | 3 μL template | for 90 sec extension at 72°C for 1 min | |
| 5'-TTGCCGATCGCGTGAAGT -3' | DNA and 0.5 μL of each primer mix | and a final extension of 10 min at 72°C11 | |
| sul2 | Sul2 F | 12.5 μL Dream Taq master mix, | 3 min at 95°C to before 35 cycles of 1 min |
| blaOXA-1 | 5'-CGGCTGCGCTTCGATT-3' | 8.5 μL nuclease-free water, | at 94°C, annealing temperature at 47°C |
| Sul2 R | 3 μL template DNA and | for 90 sec extension at 72°C for 1 min | |
| 5'CGCGCGCAGAAAGGATT-3' | 0.5 μL of each primer mix | and a final extension of 10 min at 72°C12 | |
| OXA-F | 12.5 μL Dream Taq master mix, | 3 min at 95°C before 32 cycles, with | |
| 5'-ATATCTCTACTGTTGCATCTCC-3' | 8.5 μL nuclease-free water, | 1 cycle consisting of denaturation at 94°C | |
| OXA-R | 3 μL template DNA and | for 30 sec, annealing at 54°C for 30 sec | |
| 5'-AAACCCTTCAAACCATCC-3' | 0.5 μL of each primer mix | and extension at 72oC for 1 min13 | |
| Class one | Int1 F | 12.5 μL Dream Taq master mix, | 30 sec at 95°C before 35 cycles of 30 sec |
| integron | 5'-GCCTTGCTGTTCTTCTACGG-3' | 8.5 μL nuclease-free water, | at 94°C, annealing temperature at 60°C |
| Int1 R | 3 μL template DNA and | for 2 min, extension at 72°C for 2 min | |
| 5'-GATGCCTGCTTGTTCTACGG-3' | 0.5 μL of each primer mix | and a final extension of 5 min at 72°C14 |
Electrophoresis of PCR amplicons: The PCR products (amplicons) were separated by electrophoresis on a 2% (w/v) agarose gel containing 2 μL floor safe. Electrophoresis was run at 100 V for 30 min and visualized on an itraviolet (UV) trans illuminator gel imaging system (Bio-Rad Gel Imaging system, Bio-Rad, California, USA). Bands were photographed and band positions were determined and compared to molecular weight markers (1 kb and 100 bp DNA ladders, First base, Malaysia).
Statistical analysis: Data were statistically analyzed with one-way ANOVA to determine significant differences at a 5% level of acceptance using the Statistical Product and Services Solutions Software (SPSS, version 20.0). For quality assurance purposes, the samples were spiked with a lot of internal standards to ensure satisfactory extraction and instrumental analysis to check for contamination and quality control, laboratory procedural blanks and field blanks were used.
RESULTS
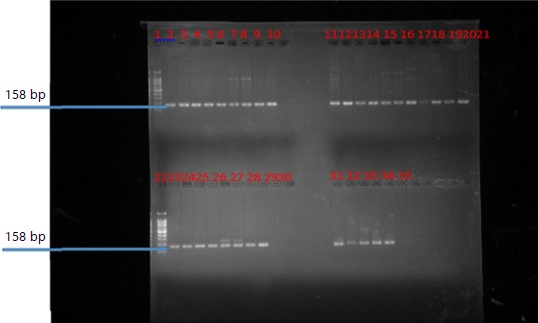
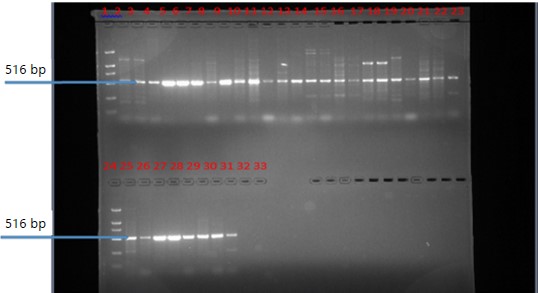
Mean values of physicochemical analysis were, the highest mean values of total nitrogen (TN) 0.72 mg L–1, phosphorus (P) 38.95 mg L–1, chemical oxygen demand (COD) 87.50 mg L–1 and total chemical oxygen demand (TCOD) 114.50 mg L–1 from samples obtained from Nigeria given in Table 2. While, total nitrogen (TN) 7.02 mg L–1, phosphorus (P) 6.79 mg L–1, chemical oxygen demand (COD) 88.50 mg L–1 and total chemical oxygen demand (TCOD) 115.50 mg L–1 were values obtained from samples from Beijing, China and given in Table 3. Solid Phase Extraction and Liquid Chromatography (SPE-LM) detected the presence of 15 out of the 17 different antibiotics assayed in the samples with Ciprofloxacin having the highest concentration shown in Table 4. Ciprofloxacin had a concentration ranging from 22.5-65.0 μg mL–1. With the highest concentration of 65.0 μg mL–1, it was the highest in the samples. Molecular characterization of antibiotic resistance genes, sulfonamide resistance genes (sul1 and sul2), a mobile gene of the class 1 integron (Int1) and the Beta lactam genes (blaTEM) all amplified from samples in the study are presented (Fig. 1-4).
| Table 2: | Physicochemical parameters of samples collected from Abuja, Nigeria | |||
| Sample | Nitrogen (mg L–1) |
Phosphorus (mg L–1) |
COD (mg L–1) |
TCOD (mg L–1) |
| 1 | 0.68±0.14 |
20.05±1.48 |
83.50±6.36 |
96.50±2.12 |
| 2 | 0.59±0.28 |
29.50±0.14 |
81.50±0.71 |
102.50±0.71 |
| 3 | 0.62±0.01 |
23.05±0.50 |
57.50±0.71 |
73.00±1.41 |
| 4 | 0.60±0.21 |
31.25±0.78 |
80.50±0.71 |
90.00±2.83 |
| 5 | 0.52±0.02 |
15.30±0.14 |
68.50±0.71 |
105.00±1.41 |
| 6 | 0.72±0.06 |
38.95±0.35 |
70.50±0.71 |
82.50±0.71 |
| 7 | 0.29±0.01 |
0.38±0.14 |
74.50±0.71 |
77.50±0.71 |
| 8 | 0.63±0.02 |
10.05±0.35 |
79.50±0.71 |
81.50±0.71 |
| 9 | 0.40±0.03 |
36.70±0.14 |
87.50±0.71 |
114.50±0.71 |
| 10 | 0.40±0.07 |
14.60±0.28 |
75.50±0.71 |
107.00±1.41 |
| ±: Standard deviation at 95% significant level | ||||
| Table 3: | Physicochemical parameters of samples collected from Beijing, China | |||
| Sample | Nitrogen (mg L–1) |
Phosphorus (mg L–1) |
COD (mg L–1) |
TCOD (mg L–1) |
| 1 | 0.52±0.01 |
4.17±0.84 |
77.00±1.41 |
96.50±0.71 |
| 2 | 0.46±0.01 |
3.22±0.11 |
82.50±0.71 |
102.00±1.41 |
| 3 | 0.22±0.01 |
0.74±0.13 |
58.00±1.41 |
70.50±0.71 |
| 4 | 0.23±0.07 |
1.69±0.05 |
83.00±2.83 |
90.00±1.41 |
| 5 | 0.34±0.02 |
1.56±0.05 |
68.00±1.41 |
105.00±1.41 |
| 6 | 0.21±0.06 |
1.88±0.04 |
74.50±0.71 |
81.50±0.71 |
| 7 | 0.20±0.01 |
1.27±0.78 |
56.50±0.71 |
84.00±2.83 |
| 8 | 3.81±0.33 |
3.40±0.13 |
73.00±1.41 |
76.50±2.12 |
| 9 | 7.02±0.28 |
1.04±0.11 |
80.00±1.41 |
80.50±2.12 |
| 10 | 0.80±0.01 |
2.85±0.01 |
82.50±0.71 |
115.50±2.12 |
| 11 | 2.11±0.01 |
0.50±0.11 |
78.50±9.19 |
105.50±0.71 |
| 12 | 0.52±0.08 |
6.79±0.09 |
73.50±2.12 |
105.00±1.41 |
| 13 | 0.86±0.06 |
4.45±0.84 |
76.50±0.71 |
101.50±0.71 |
| 14 | 1.51±0.06 |
3.33±0.01 |
86.50±0.71 |
89.50±0.71 |
| 15 | 4.09±0.11 |
3.19±0.09 |
81.50±0.71 |
92.00±1.41 |
| 16 | 1.06±0.23 |
1.47±0.06 |
83.00±2.83 |
83.00±1.41 |
| 17 | 0.15±0.01 |
0.23±0.03 |
88.50±2.12 |
84.50±0.71 |
| 18 | 2.00±0.16 |
0.35±0.01 |
78.50±0.71 |
85.00±1.41 |
| 19 | 1.55±0.04 |
1.05±0.67 |
84.50±0.71 |
86.50±0.71 |
| 20 | 1.82±0.10 |
1.07±0.25 |
68.00±1.41 |
89.00±1.41 |
| 21 | 0.55±0.04 |
0.57±0.63 |
57.00±1.41 |
64.50±0.71 |
| 22 | 1.50±0.21 |
1.30±0.07 |
76.50±0.71 |
74.00±1.41 |
| ±: Standard deviation at 95% significant level | ||||
| Table 4: | Mean concentration of antibiotics in samples | |||
| Ampicilin | 5.7 |
0 |
0 |
0 |
0 |
0 |
5.5 |
5.4 |
0 |
5.5 |
0 |
0 |
0 |
5.5 |
| Ciprofloxacin | 65 |
47.1 |
48.5 |
45.5 |
44.5 |
46.8 |
48.2 |
48.6 |
45.7 |
46.1 |
44.2 |
49.6 |
45.7 |
22.5 |
| Chlorotetracycline | 3.5 |
2.4 |
3.01 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2.93 |
| Cefotaxime | 0 |
0 |
1.8 |
0 |
0 |
2.2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Enrofloxacin | 42.3 |
41.2 |
38.8 |
38 |
39 |
37.7 |
38.3 |
38.2 |
37.9 |
37.8 |
38 |
37.6 |
38.1 |
38.3 |
| 4-Eploxytetracycline | 25.2 |
0 |
41.7 |
19.4 |
25.2 |
0 |
14.9 |
19 |
39.2 |
16.4 |
37 |
37.6 |
34.8 |
33.8 |
| 4-epitetracycline hydrochloride | 13 |
13 |
14.2 |
12.9 |
14.2 |
12.7 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Sulfamethiazole | 2.2 |
2 |
2 |
4.1 |
4.1 |
1.1 |
0 |
4.3 |
2 |
1.9 |
2.1 |
5 |
4.1 |
4.5 |
| Sulfadiazine | 10.8 |
11.9 |
24 |
12.1 |
24.3 |
13.3 |
11.5 |
26 |
0 |
12.4 |
11.2 |
12.9 |
23.9 |
11.4 |
| Sulfameracine | 0 |
1.2 |
0.8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Sulfamonomethodine sodium | 5 |
5.4 |
4.1 |
2.7 |
2.4 |
2.4 |
3 |
2.2 |
5.3 |
3.5 |
0 |
0 |
2.6 |
0 |
| Roxithromycin | 0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Norfloxacin | 44.9 |
45 |
42.1 |
41 |
42.1 |
39.6 |
40.6 |
42.4 |
40.4 |
39.7 |
40.2 |
41.6 |
19.9 |
41.2 |
| Ofloxadin | 46.2 |
34.2 |
35 |
35.3 |
33.4 |
34 |
33.2 |
33.6 |
34.5 |
33.5 |
34 |
33.7 |
33.4 |
16.7 |
| Oxytetracycline | 10.2 |
10.2 |
20.5 |
10.1 |
0 |
0 |
10.2 |
0 |
0 |
0 |
10.8 |
10.2 |
10.2 |
10.2 |
| Penicilin | 8.9 |
11.5 |
2.4 |
4.8 |
4.6 |
4.7 |
9.4 |
10.8 |
25.3 |
5.4 |
11.8 |
4 |
3 |
5.5 |
| Tetracyclines | 21.6 |
10.7 |
10.9 |
21.1 |
21.9 |
11 |
0 |
10.9 |
0 |
0 |
0 |
0 |
0 |
22 |
| Samples (mg L–1) | 51 |
52 |
53 |
54 |
55 |
56 |
57 |
58 |
59 |
510 |
511 |
512 |
513 |
514 |
|

|

|
|
DISCUSSION
The present study revealed high levels of some physicochemical parameters which represent potential public health risks. The concentration of phosphates exceeded the acceptable WHO limits of 3.5, 15 and 10 mg L–1, respectively in most samples from both study areas. The excessive presence of phosphate in conjunction with nitrates and potassium has been reported to cause algal blooms as phosphate is the nutrient source for algae15. Eutrophication intensification is associated with an increase of nutrient anthropogenic loads consisting mainly of phosphorus and nitrogen which are discharged into surface water and other receiving water bodies16. Although the concentration of Nitrogen in samples in this study was below the standard limits of the nitrate quality guideline established by Canadian environmental quality guidelines for the protection of aquatic life (CCME) of 13 mg L–1. Monitoring is important to maintain acceptable concentrations. Chemical oxygen demand (COD) is the measure of oxygen equivalent of the organic content of a sample that is susceptible to oxidation by a strong chemical oxidant. It is an evaluation used to measure the level of water contamination by organic matter17. The COD of the water samples analyzed from both study areas exceeded the limits for effluent discharge of 30, 75 and 150 mg L–1, respectively. The presence of oxidizable inorganic compounds may have been responsible for the high concentration of chemical oxygen demand in samples and indicates high contamination. The values of physicochemical parameters obtained in our study are indicators of poor water quality, hence, potential risks to human, animal and environmental health. The result of this study is in tandem with reports that the physicochemical and microbial quality of freshwater bodies is under great stress due to the gulping of huge amounts of waste18.
Polymerase chain reaction assayed and amplified the following antibiotic resistance genes from our samples, sulfonamide resistance genes (sul1 and sul2), mobile genetic element, Class1 integron (int1) and Beta-lactam (blaOXA-1) genes. The amplified resistance genes in the study are a reflection of the abundance of antibiotics in the studied environment. The aquatic environment has been reported to be critical in the propagation of antimicrobial resistance determinants in the environment as they aid dissemination19. The int1, one of the gene amplified, is a mobile genetic element and has been reported to capture, integrate and express resistance gene cassettes in their variable regions thereby transferring resistance horizontally to other bacteria and microorganisms20, making nonresistant organisms resistant. The int1 gene has previously been identified as a measurement of horizontal genetic transmission (HGT) potential and gene acquisition21 and it has been reported as an indicator of anthropogenic pollution22. This further confirms the very high use and misuse of antibiotics in the study areas. The amplification of sul1 and sul2 genes in both study areas represents a potential public health risk as both genes have been associated with treatment failure23. The potential risks of these specific genes would be exacerbated by the possibility to be transferred horizontally between strains24. The amplification of the blaOXA-1 genes also represents potential risks and is not surprising as β-lactamase antibiotics have been reported as one of the most prescribed drugs worldwide5. Problematic and hard-to-treat infections are reported to be caused by β-lactamase (ESBL)-producing Enterobacteriaceae24. Resistance to β-lactamase antibiotics indicates possible treatment failure and could be life-threatening, hence a high risk to public health. Earlier reports from a study of regional environmental distribution simulations and risk assessments for commonly used fluoroquinolones in the Wenyu River catchment and upstream Shahe Reservoir reported the presence of high Ciprofloxacin25. Interestingly, the Wenyu River was part of the study locations in this research and with similar results reflecting antibiotics and antibiotics resistance genes abundance.
Other studies in China associated the increasingly high usage of antibiotics in human and animal therapy with its corresponding impact on the environment/water bodies26. In Nigeria, the increase in antibiotic usage has been attributed to poor regulatory control of antibiotic sales and purchases without prescription27. One of the fluoroquinolones assayed for in the present study, Ciprofloxacin had the highest detection level in all the samples (from both study areas). The presence of these resistance genes shows the risk involved in improper wastewater treatment and reuse. Ciprofloxacin, are among the high antibiotics of last resort and resistance to it indicates, antibiotics misuse and calls for concern.
With continuous population growth and the additional crisis of increased demand for water and diminishing freshwater resources, wastewater reclamation for agricultural, industrial and other purposes has become a necessity. However, it requires adequate and effective treatment before reuse as improperly treated wastewater represents environmental pools for antibiotic-resistant bacteria and antibiotic-resistant genes.
Wastewater through a resource, requires proper monitoring for emerging contaminants and adequate treatment before release, reuse or disposal in the environment is important. Research for effective and low-cost wastewater treatment and recovery processes as a water action towards the achievement of the sustainable development goals especially goal 6 is recommended.
CONCLUSION
The study confirmed the presence of targeted contaminants and revealed phenotypic and molecular evidence of antibiotics and antibiotic-resistance genes in samples obtained from both regions. Wastewater treatment research which has the potential to advance safe, reliable and cost-effective technologies to reuse effluents should be a national and international priority.
SIGNIFICANCE STATEMENT
The study assessed the presence of residues of selected commonly used antibiotics and antibiotic-resistance genes in wastewater from two treatment plants. Molecular characterization confirmed the presence of antibiotic resistance genes in samples (blaTEM, 526 bp, sul1, 158 bp, sul2, 158 bp and int, 473 bp), revealing evidence of antibiotics and antibiotic resistance genes. The paper highlights the significance of the rational use of wastewater and opines that a high premium is placed on research on wastewater treatment and utilization as it has the potential to advance safe, reliable and cost-effective technologies to reuse.
ACKNOWLEDGMENTS
The facilities provided at the Research Centre for Eco-Environmental Sciences, Chinese Academy of Sciences Beijing, China is gratefully acknowledged. The technical support during sample collections at both study areas is thankfully acknowledged.
REFERENCES
- Mbanga, J., A.L.K. Abia, D.G. Amoako and S.Y. Essack, 2020. Quantitative microbial risk assessment for waterborne pathogens in a wastewater treatment plant and its receiving surface water body. BMC Microbiol., 20: 364.
- Angelakis, A.N. and S.A. Snyder, 2015. Wastewater treatment and reuse: Past, present, and future. Water, 7: 4887-4895.
- Liu, N., W. Jiang, L. Huang, Y. Li, C. Zhang, X. Xiao and Y. Huang, 2022. Evolution of sustainable water resource utilization in Hunan Province, China. Water, 14: 2477.
- Han, H., M. Bai, Y. Chen, Y. Gong and M. Wu et al., 2021. Dynamics of diversity and abundance of sulfonamide resistant bacteria in a silt loam soil fertilized by compost. Antibiotics, 10: 699.
- Teklu, D.S., A.A. Negeri, M.H. Legese, T.L. Bedada, H.K. Woldemariam and K.D. Tullu, 2019. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control, 8: 39.
- Pärnänen, K.M.M., C. Narciso-da-Rocha, D. Kneis, T.U. Berendonk and D. Cacace et al., 2019. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv., 5.
- Zhang, Q.Q., G.G. Ying, C.G. Pan, Y.S. Liu and J.L. Zhao, 2015. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol., 49: 6772-6782.
- Abakpa, G.O., V.J. Umoh, S. Kamaruzaman and M. Ibekwe, 2018. Fingerprints of resistant Escherichia coli O157:H7 from vegetables and environmental samples. J. Sci. Food Agric., 98: 80-86.
- Becerra-Castro, C., A.R. Lopes, I. Vaz-Moreira, E.F. Silva, C.M. Manaia and O.C. Nunes, 2015. Wastewater reuse in irrigation: A microbiological perspective on implications in soil fertility and human and environmental health. Environ. Int., 75: 117-135.
- Pernet-Coudrier, B., W. Qi, H. Liu, B. Muller and M. Berg, 2012. Sources and pathways of nutrients in the semi-arid region of Beijing-Tianjin, China. Environ. Sci. Technol., 46: 5294-5301.
- Heuer, H. and K. Smalla, 2007. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol., 9: 657-666.
- Heuer, H., A. Focks, M. Lamshöft, K. Smalla, M. Matthies and M. Spiteller, 2008. Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol. Biochem., 40: 1892-1900.
- Colom, K., J. Pérez, R. Alonso, A. Fernández-Aranguiz, E. Lariño and R. Cisterna, 2003. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett., 223: 147-151.
- Shahcheraghi, F., F. Badmasti and M.M. Feizabadi, 2010. Molecular characterization of class 1 integrons in MDR Pseudomonas aeruginosa isolated from clinical settings in Iran, Tehran. FEMS Immunol. Med. Microbiol., 58: 421-425.
- Bhateria, R. and D. Jain, 2016. Water quality assessment of lake water: A review. Sustainable Water Resour. Manage., 2: 161-173.
- Preisner, M., E. Neverova-Dziopak and Z. Kowalewski, 2021. Mitigation of eutrophication caused by wastewater discharge: A simulation-based approach. Ambio, 50: 413-424.
- Alrumman, S.A., A.F. El-kott And S.M.A.S. Keshk, 2016. Water pollution: Source & treatment. Am. J. Environ. Eng., 6: 88-98.
- Haque, M.A., M.A.S. Jewel and M.P. Sultana, 2019. Assessment of physicochemical and bacteriological parameters in surface water of Padma River, Bangladesh. Appl. Water Sci., 9: 10.
- Milobedzka, A., C. Ferreira, I. Vaz-Moreira, D. Calderon-Franco and A. Gorecki et al., 2022. Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the One-Health cycle. J. Hazard. Mater., 424: 127407.
- Gundogdu, A., Y.B. Long, T.L. Vollmerhausen and M. Katouli, 2011. Antimicrobial resistance and distribution of sul genes and integron-associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J. Med. Microbiol., 60: 1633-1642.
- Narciso-da-Rocha, C., A.R. Varela, T. Schwartz, O.C. Nunes and C.M. Manaia, 2014. blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital-urban wastewater treatment plant system. J. Global Antimicrob. Resist., 2: 309-315.
- Paulus, G.K., L.M. Hornstra, N. Alygizakis, J. Slobodnik, N. Thomaidis and G. Medema, 2019. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health, 222: 635-644.
- Gillings, M.R., 2017. Class 1 integrons as invasive species. Curr. Opin. Microb., 38: 10-15.
- Lerminiaux, N.A. and A.D.S. Cameron, 2019. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol., 65: 34-44.
- Hao, X., Y. Cao, L. Zhang, Y. Zhang and J. Liu, 2015. Fluoroquinolones in the Wenyu River catchment, China: Occurrence simulation and risk assessment. Environ. Toxicol. Chem., 34: 2764-2770.
- Shen, L., X. Wei, J. Yin, D.R. Haley, Q. Sun and C.S. Lundborg, 2022. Interventions to optimize the use of antibiotics in China: A scoping review of evidence from humans, animals, and the environment from a one health perspective. One Health, 14: 100388.
- Ajibola, O., O.A. Omisakin, A.A. Eze and S.A. Omoleke, 2018. Self-Medication with antibiotics, attitude and knowledge of antibiotic resistance among community residents and undergraduate students in Northwest Nigeria. Diseases, 6: 32.
How to Cite this paper?
APA-7 Style
Abakpa,
G.O., Wei,
Y., Obioh,
G., John,
U.A. (2023). Wastewater Treatment Plants; Hot Spots of Emerging Contaminants in the Environment. Research Journal of Microbiology, 18(1), 11-19. https://doi.org/10.3923/rjm.2023.11.19
ACS Style
Abakpa,
G.O.; Wei,
Y.; Obioh,
G.; John,
U.A. Wastewater Treatment Plants; Hot Spots of Emerging Contaminants in the Environment. Res. J. Microbiol 2023, 18, 11-19. https://doi.org/10.3923/rjm.2023.11.19
AMA Style
Abakpa
GO, Wei
Y, Obioh
G, John
UA. Wastewater Treatment Plants; Hot Spots of Emerging Contaminants in the Environment. Research Journal of Microbiology. 2023; 18(1): 11-19. https://doi.org/10.3923/rjm.2023.11.19
Chicago/Turabian Style
Abakpa, Grace, Onyukwo, Yausong Wei, Gloria Obioh, and Ujoh Adole John.
2023. "Wastewater Treatment Plants; Hot Spots of Emerging Contaminants in the Environment" Research Journal of Microbiology 18, no. 1: 11-19. https://doi.org/10.3923/rjm.2023.11.19

This work is licensed under a Creative Commons Attribution 4.0 International License.



